
Since its inception within the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center (GMTEC) in 2017, SAIL has grown to serve more than 80 laboratories spanning basic and translational research. Our mission is to assimilate emerging technologies in the rapidly advancing fields of single-cell sequencing and imaging-based profiling, in addition to providing robust analytical solutions.
Because MSK has incomparable resources: unparalleled clinical volume and access to human samples from both the clinic and research autopsies; preeminent clinical expertise; a wealth of experimental tools, including mouse and organoid models; and computational leadership, SAIL technologies have been applied to a spectrum of the most pressing basic and translational research questions at MSK, including the study of basic developmental processes, metastasis, immunotherapy, and wound healing. The SAIL team is continually innovating new protocols to access extremely challenging clinical tissues and is involved in basic technological development. We participate in high-profile projects, such as the study of lung and pancreatic cancer metastasis as part of the Human Tumor Atlas Network of the National Cancer Institute.
SAIL is constantly comparing and testing different samples preparation methods or single-cell / spatial genomics approaches. Amongst our most recent efforts we have:
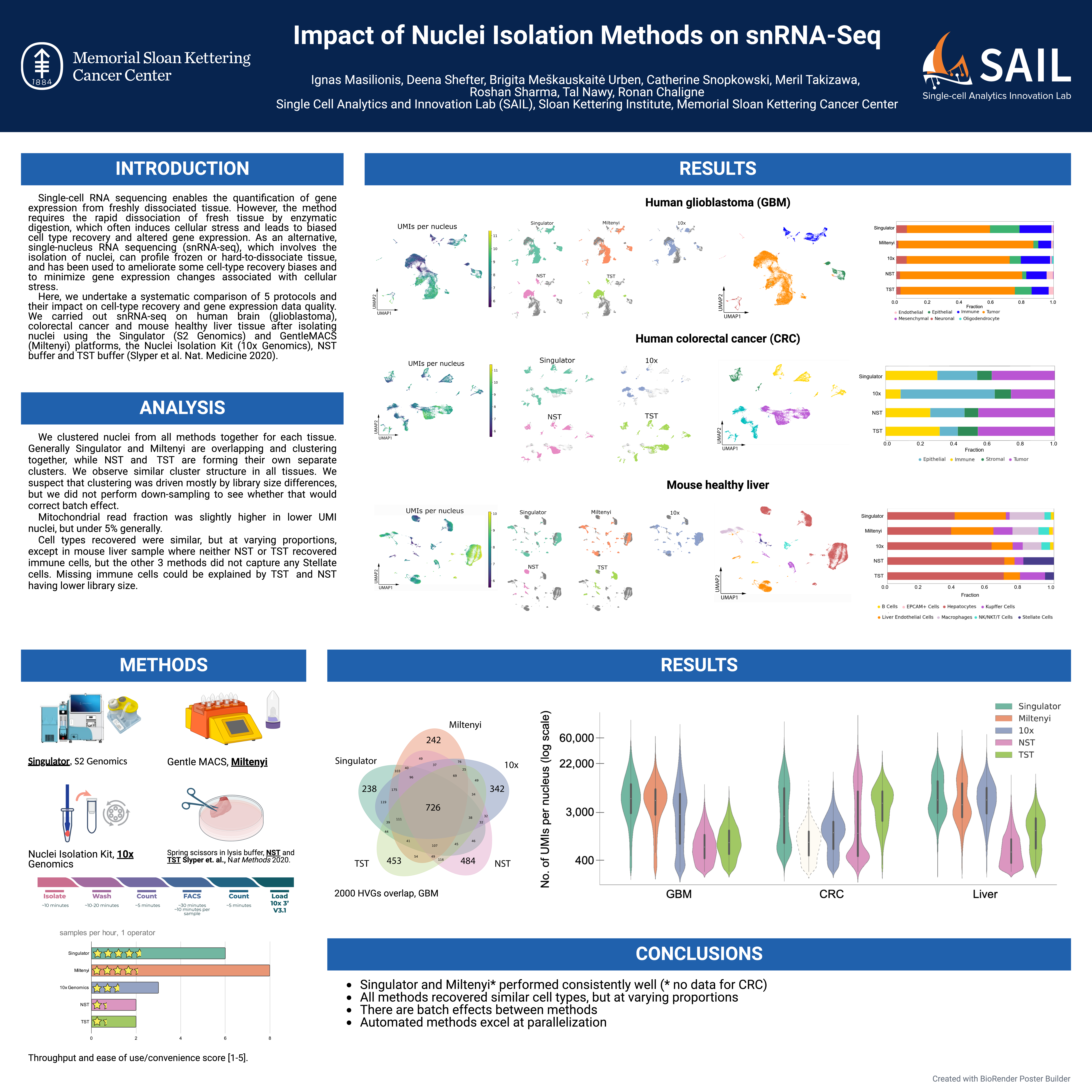
Conducted comprehensive benchmarking studies comparing nuclei preparation techniques (Singulator by S2 Genomics, Miltenyi, 10X Genomics, and previously published method, Slyder et al), has shown that Singulator and Miltenyi consistently out-performed other methods on the three different tissues (based on UMIs count and cell types recovered) we have tested (CRC, Healthy mouse liver and GBM). To disseminate these findings, we are currently preparing a manuscript and have been presenting those results at conference (Keystone in June 2023, Human Cell Atlas in July 2023).
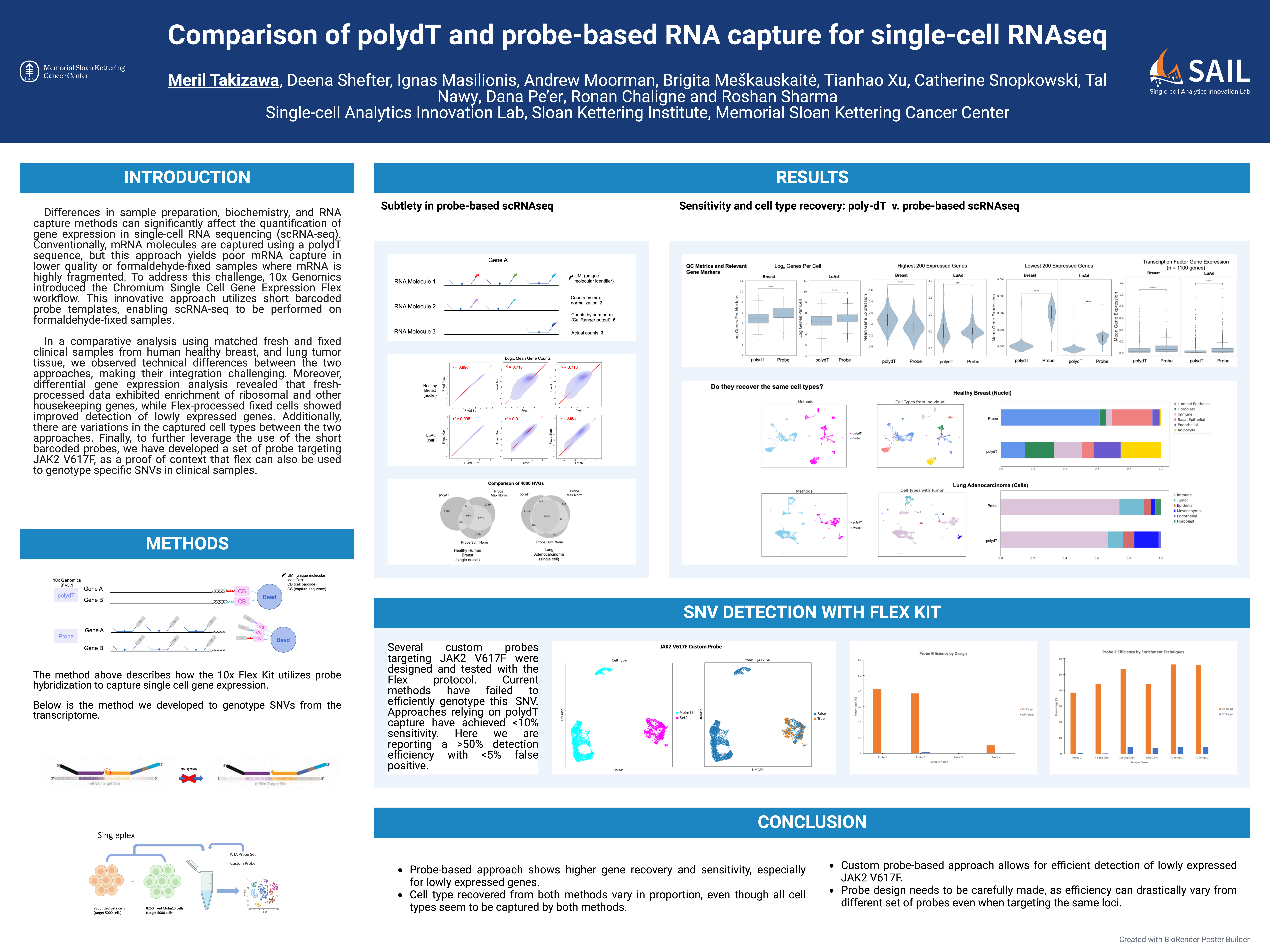
Evaluated fixed (Flex kit) versus fresh samples (3’ v3.1) methods by 10X Genomics on two different challenging sample types (LUAD and Healthy breast tissue) presenting our results at multiple conferences to inform the scientific community and improve experimental practices.
Conducted head-to-head comparisons of five different dead cell removal protocols, including LeviCell, FACS, Miltenyi, Akadeum, and Stem Cell, assessing what methods is the least “traumatic” for fragile cell type. This comparison has been conducted on CSF (Boire’s lab) and Lymph Node tissue (Brown’s lab), which have been two tissue showing stress during sample preparation in the past.
Those results have been and will be presented at various conferences this year. We are hoping to have those published online really soon too. Please reach out to us if you want more details.
Over the years, we have extensively optimized sample preparation protocols for a wide array of mouse and human tissues. Our expertise ranges from straightforward samples like blood to more complex specimens such as pancreas and prostate.
SAIL has successfully processed tumor biospecimens obtained from clinical resections, core needle biopsies, fine needle aspirates, and liquid sources. We have also invested significant effort into optimizing multi-omics approaches, such as combining single-cell RNA sequencing (scRNAseq) with single-cell ATAC sequencing (scATACseq). Through extensive collaboration, including our participation in the Human Tumor Atlas Network, we have demonstrated the feasibility of generating robust single-cell multi-omics datasets.
For instance, we have recently:
Implemented DOGMAseq, an innovative multiomics method developed at NYGC, facilitating native lineage tracing through mitochondrial DNA sampling, along with achieving higher gene expression recovery compared to the traditional 10X Genomics Multiome methods.
Partnered with S2 Genomics, utilizing their Singulator instrument to enhance throughput and consistency in nuclei preparation. Over the past year, more than 400 samples have been processed with the Singulator at SAIL for snRNAseq, snATAC, Multiome, DLP+ and Mission Bio Tapestry.
Implemented and optimized protocols to apply snRNAseq flex approach to FFPE samples, broadening the scope of single-cell analysis to include archived and fixed tissues.
Improved enrichment for fragile cell type using LeviCell EOS by LevitasBio
SAIL has implemented Visium FFPE and CytAssist workflow, significantly enhancing sensitivity and efficiency in processing FFPE samples, broadening the scope of spatial transcriptomics studies.
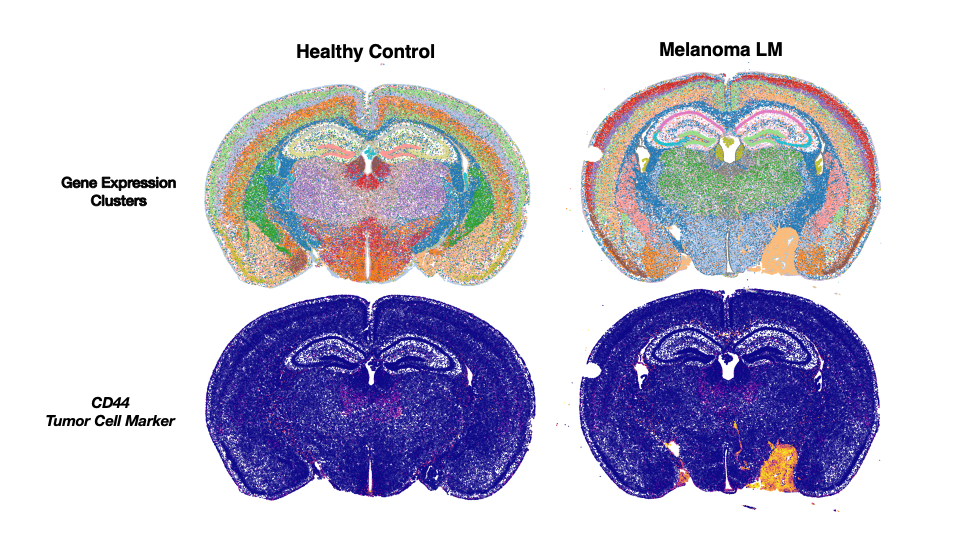
We also have purchased and implemented Xenium In Situ, a spatial transcriptomics solution enabling the mapping of gene expression patterns within tissue samples, expanding our spatial analysis capabilities.